Привіт Гість ( Вхід | Реєстрація )
| nikelong |
 Jan 2 2010, 02:07 Jan 2 2010, 02:07
Пост
#1
|
|
Тера ранчер           Група: Trusted Members Повідомлень: 11 909 З нами з: 19-March 05 Користувач №: 92 Стать: Чол |
 Проект "RNA World (beta)" Проект "RNA World (beta)"----------------------------------------------------------------------------------------------------------
ТОП-20 участников:  ---------------------------------------------------------------------------------------------------------- Дата основания команды - 03.01.2010 Капитан - distributed.org.ua ---------------------------------------------------------------------------------------------------------- Для присоединения к команде Украины: 1. Загрузите BOINC менеджер (Если его у Вас еще нет!) 2. Перейдите в "расширенный вид" 3. Выберите сервис ---> добавить проект 4. Введите адрес проекта http://www.rnaworld.de/rnaworld/ 5. Введите свои регистрационные данные. 6. Найдите нашу команду. Она называется Ukraine. Нажмите Join чтобы вступить в команду. 7. Если есть доступные для загрузки задания Вы их получите и начнете расчеты. ---------------------------------------------------------------------------------------------------------- Новичкам: статья со скриншотами, как поставить и настроить BOINC-менеджер ---------------------------------------------------------------------------------------------------------- Полезная информация: Для идентификации пользователя в BOINC могут служить 2 вещи: 1) пара e-mail/пароль 2) межпроектный идентификационный ID (Cross-project ID) - 32значное шестнадцатиричное число. Если Вы пожелаете подключится ещё и к другому BOINC-проекту, то помните: чтобы не плодить новых аккаунтов при подключении к новому проекту или команде, нужно обязательно везде регистрироваться с одним и тем же Именем и EMAIL. если при регистрации в проекте указать другой e-mail , BOINC создаст новый аккаунт с тем же именем! В этом случае рекомендуется зайти во все ваши аккаунты и во все проекты и где надо поменять емейл на нужный. Через некоторое время ваши аккаунты сольются в один с одним cross-project-id. ---------------------------------------------------------------------------------------------------------- О проекте - общая информация (на английском): RNA World занимается исследованием РНК разных оргинизмов: от простейших, до РНК человека (Show/Hide) Подпроекты: (ссылка на официальную страницу) Project: CRISPR (Show/Hide) Project: GEMM (Show/Hide) Project: 6S (Show/Hide) Project: Thermo (Show/Hide) Project: Mtb, Myctu, Mycle & Myc - изучает РНК туберкулезных палочек (Show/Hide) Project: sRib (Show/Hide) Project: tRNA & tRNA-like (Show/Hide) Project: T-box (Show/Hide) Project: GA (Show/Hide) ------------------------- Ссылки по теме:
(Show/Hide) Це повідомлення відредагував nikelong: Sep 18 2010, 20:35 |
  |
Відповідей(105 - 119)
| Salmonella |
 Jun 18 2010, 11:04 Jun 18 2010, 11:04
Пост
#106
|
|
кранчер-новачок    Група: Trusted Members Повідомлень: 38 З нами з: 10-June 10 Користувач №: 1 414 Стать: Чол |
RNA World conference presentation Today I would like to inform you that we will present the RNA World distributed computing project at the RNA 2010 conference of the RNA Society held next week in Seattle/USA. Thanks a lot for your valuable contributions which ultimately made this presentation possible and hopefully will allow us to make new contacts to many RNA researchers in order to establish additional cooperation projects. Please also note that this will be the second official presentation of the RNA World project within eight months, so we are quite happy about that and will now prepare to publish our first scientific results in a peer-reviewed journal. Michael. 17 Jun 2010 22:45:26 UTC Сегодня я хотел бы сообщить вам, что мы представляем проект распределенных вычислений на RNA 2010 http://depts.washington.edu/rna2010/ конференции RNA Society http://www.rnasociety.org/ состоится на следующей неделе в Сиэтле / США. Большое спасибо за Ваш ценный вклад которых в конечном итоге сделал эту презентацию возможной, и мы надеемся, позволит нам сделать новые контакты со многими исследователями РНК в целях создания дополнительных проектов сотрудничества. Пожалуйста, обратите внимание, что это будет вторая официальная презентация проекта RNA World в течение восьми месяцев, так что мы очень рады, что и сейчас готовятся к публикации наши первые научные результаты в специальном журнале. Как-то так, примерно. |
| Rilian |
 Jun 26 2010, 16:39 Jun 26 2010, 16:39
Пост
#107
|
 interstellar           Група: Team member Повідомлень: 17 163 З нами з: 22-February 06 З: Торонто Користувач №: 184 Стать: НеСкажу Free-DC_CPID Парк машин: ноут и кусок сервера |
2010-06-25: Just a short note on the latest developments: I am currently experiencing remarkable interest in our project at the RNA conference in Seattle. It seems that quite a number of people is interested in cooperating with us in the future. Some of these people do have full trancriptome sequences available that would help us to validate our computational results on a lab experimental basis. So, I think these are really exciting news.
Michael. -------------------- |
| Salmonella |
 Jun 28 2010, 03:11 Jun 28 2010, 03:11
Пост
#108
|
|
кранчер-новачок    Група: Trusted Members Повідомлень: 38 З нами з: 10-June 10 Користувач №: 1 414 Стать: Чол |
Решил перевести страничку "Описание проекта". Вот что получилось.
http://www.rechenkraft.net/wiki/index.php?...tdescription/ru |
| nikelong |
 Jun 28 2010, 21:26 Jun 28 2010, 21:26
Пост
#109
|
|
Тера ранчер           Група: Trusted Members Повідомлень: 11 909 З нами з: 19-March 05 Користувач №: 92 Стать: Чол |
Salmonella,
Отличная работа! Но...может быть вскользь - но упомянуть наш сайт или линк на эту тему на той странице? Чтобы людей из Украины соорентировать... -------------------- |
| Salmonella |
 Jul 1 2010, 05:17 Jul 1 2010, 05:17
Пост
#110
|
|
кранчер-новачок    Група: Trusted Members Повідомлень: 38 З нами з: 10-June 10 Користувач №: 1 414 Стать: Чол |
Salmonella, Отличная работа! Но...может быть вскользь - но упомянуть наш сайт или линк на эту тему на той странице? Чтобы людей из Украины соорентировать... Даже не знаю. Это надо с yoyo говорить. Поляки и на отдельной странице дополнительно сделали, например. http://www.boincatpoland.org/wiki/RNAWorld |
| ReMMeR |
 Jul 7 2010, 10:13 Jul 7 2010, 10:13
Пост
#111
|
 ----===[ oO ]===----          Група: Team member Повідомлень: 2 910 З нами з: 20-October 05 З: Quake arena Користувач №: 135 Стать: Чол Free-DC_CPID |
Реквестирую в шапку инструкцию как подрубиться по слабому ключу на коммандный аккаунт distributed.org.ua (если это возможно).
П.с в этом проекте компндный аккаунт не на том кросс-проджект ай-ди, что в остальных. -------------------- (Show/Hide) |
| Rilian |
 Aug 16 2010, 02:25 Aug 16 2010, 02:25
Пост
#112
|
 interstellar           Група: Team member Повідомлень: 17 163 З нами з: 22-February 06 З: Торонто Користувач №: 184 Стать: НеСкажу Free-DC_CPID Парк машин: ноут и кусок сервера |
As announced, the second Rfam 10.0-based CMCALIBRATE WU set has been uploaded to the server and will be processed as soon as the current WU archive generation (CMSEARCH) has completed.
-------------------- |
| nikelong |
 Sep 13 2010, 09:21 Sep 13 2010, 09:21
Пост
#113
|
|
Тера ранчер           Група: Trusted Members Повідомлень: 11 909 З нами з: 19-March 05 Користувач №: 92 Стать: Чол |
Бактерии открыли окно в прошлое
16.03.2009 Некоторые бактерии умеют плавать, видоизменяться и переносить вирусы – и все эти способности исправно работают, несмотря на отсутствие у этих микроорганизмов ДНК. Исследование такого вида одноклеточных существ дает ученым возможность узнать, как выглядели первые представители органической жизни на Земле. Теория о том, что при своем зарождении организмы не имели ДНК, находит множественные подтверждения в ходе различных исследований. Для обозначения подобных форм существования в лексиконе ученых давно появился термин «РНК-мир». Микробиологи из Йельского университета изучили, как бактерии обретают свои необычные свойства, и это знание позволит заглянуть в далекое прошлое и представить себе ранние формы жизни. Оказалось, что одноклеточные могут активизировать свои навыки при помощи РНК, которые принято считать побочным набором кислот, интерпретирующим команды ДНК и создающим протеины. Конечно, протеины исключительно важны для функционирования любого организма, однако функции РНК тем не ограничиваются. Эти кислоты также играют неожиданно большую роль во всех проявлениях клеточной активности. Исследования американских ученых наглядно продемонстрировали, что не всегда для работы фундаментальных способностей клеток нужны протеины. Руководитель проекта профессор Рональд Брикер уверен, что подобный внутриклеточный механизм был свойственен и обитателям древнего мира. Версия для печати Шрифт Послать другу Главным результатом работы Брикера и его подопечных стало описание процесса переключения молекул РНК, известных под названием цикличный дигуанилейт (di-GMP). Эти крошечные компоненты РНК отвечают за включение и выключение работы тех или иных генов, и именно их деятельностью обусловлены умения бактерий плавать, застывать на месте и объединяться с другими одноклеточными в симбиотичный организм – биофильм. Цикличные дигуанилейты состоят всего из двух мельчайших компонентов РНК – нуклеотидов, однако способны запускать огромную (по клеточным меркам) структуру – рибопереключатель. Этот элемент РНК обычно занимается тем, что, получив инструкции от ДНК, определяет – какие гены включить. В случае бактерий, когда нет источника команд, рибопереключатель становится объектом манипуляций дигуанилейтов. Полученные результаты исследования помогут ученым узнать больше о структуре клеток и об их функционировании. Кроме того, на основании полученных данных, возможно, получится воссоздать картину зарождения органической жизни на Земле. Сам Брикер сравнил свои открытия с поисками Эльдорадо. «Мы представляли, что где-то в этих джунглях находится затерянный РНК-город. И мы просто пошли в нужном направлении и нашли его», - говорит ученый. http://www.pravda.ru/science/eureka/discov...760-rnaworld-0/ -------------------- |
| nikelong |
 Sep 19 2010, 08:43 Sep 19 2010, 08:43
Пост
#114
|
|
Тера ранчер           Група: Trusted Members Повідомлень: 11 909 З нами з: 19-March 05 Користувач №: 92 Стать: Чол |
2010-09-19: After the RNA World project presentation at the RNA 2010 conference in June in Seattle/USA and a seminar talk given at the Massachusetts Institute of Technology in Cambridge/USA on 9th of September, the RNA World project team currently presents the RNA World distributed supercomputer project outline and some of the acquired results at the 126th conference of the Gesellschaft Deutscher Naturforscher und Г„rzte e. V. (GDNГ„) in Dresden/Germany (Twitter tweets).
Michael. http://www.rnaworld.de/rnaworld/forum_thread.php?id=65 -------------------- |
| corsar83 |
 Oct 14 2010, 11:24 Oct 14 2010, 11:24
Пост
#115
|
|
кранчер зі стажем       Група: Trusted Members Повідомлень: 426 З нами з: 28-January 10 Користувач №: 1 282 Стать: Чол Free-DC_CPID Парк машин: Pentium E6500 2,93Ghz, Phenom X6 1055T 2.8Ghz Gigabyte 5870OC(875/4800), Radeon 5750 |
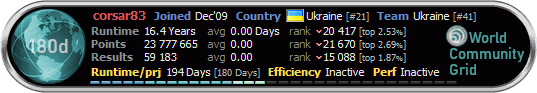
New binaries for Windows machines
As of today, new binaries for CMSEARCH have been released for Windows 32/64 bit systems. These should solve a bug which was identified with a very demanding subset of WUs. Our tests have shown that this bug should no longer occur. New binaries for Linux machines will be released soon as well. We are currently testing some additional speed improvements for Linux using the Intel compiler.[indent] New binaries for Linux In the meantime we have also replaced the Linux CMSEARCH x32/64 binaries. In a few days, we intend to again change all binaries to add another approx. 10% performance improvement by usage of the Intel C++ Compiler (yes, AMD CPUs will profit the same way as Intel CPUs do). Michael. 9 Oct 2010 16:33:44 UTC · Комментарий -------------------- |
| tiss |
 Nov 4 2010, 10:25 Nov 4 2010, 10:25
Пост
#116
|
 Мега ранчер         Група: Trusted Members Повідомлень: 1 640 З нами з: 28-February 09 Користувач №: 952 Стать: Чол Free-DC_CPID |
Ваще офигели cms_GA[e30-50MB_Lin64s]_Oryzias-latipes-(Japanese-medaka)_DG000017.lin.EMBL_RF00640_MIR167_1_1284912637_49484
Completed and validated Runtime 378,386.23 Granted credit 2,294.95 ППц 4 суток считать хрень какую-то... --------------------  |
| Rilian |
 Nov 4 2010, 11:43 Nov 4 2010, 11:43
Пост
#117
|
 interstellar           Група: Team member Повідомлень: 17 163 З нами з: 22-February 06 З: Торонто Користувач №: 184 Стать: НеСкажу Free-DC_CPID Парк машин: ноут и кусок сервера |
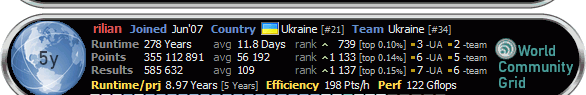
tiss, 22 очка в час, довольно неплохо...
-------------------- |
| Rilian |
 Mar 17 2011, 19:36 Mar 17 2011, 19:36
Пост
#118
|
 interstellar           Група: Team member Повідомлень: 17 163 З нами з: 22-February 06 З: Торонто Користувач №: 184 Стать: НеСкажу Free-DC_CPID Парк машин: ноут и кусок сервера |
RNA World: Reorganization of the CMSEARCH application
2011-03-14: We are currently working on reorganizing the CMSEARCH application into two variants of which one will run a combined package of many very small WUs while the other runs the rest (the default CMSEARCH) WUs. With this we intend to achieve three goals. First, the combination of many tiny WUs into one package tremendously reduces the client-server communication load: a big relieve for our server which will make it significantly more stable even during team challenges and races. Second, the packaged WUs will write a checkpoint after each completed sub-WU. Third, because the users will be allowed to opt in or out for each of these two CMSEARCH variants there will be much more flexibility at the user's end to control maximum CMSEARCH runtimes -------------------- |
| Salmonella |
 Jun 9 2011, 06:47 Jun 9 2011, 06:47
Пост
#119
|
|
кранчер-новачок    Група: Trusted Members Повідомлень: 38 З нами з: 10-June 10 Користувач №: 1 414 Стать: Чол |
|
| Rilian |
 Sep 17 2011, 21:45 Sep 17 2011, 21:45
Пост
#120
|
 interstellar           Група: Team member Повідомлень: 17 163 З нами з: 22-February 06 З: Торонто Користувач №: 184 Стать: НеСкажу Free-DC_CPID Парк машин: ноут и кусок сервера |
Помогите команде Украины стать обратно на 9 место в DC-Vault -- в RNA мы на 48 месте о_О!
http://dc-vault.com/showteam.php?team=433 -------------------- |
  |
1 Користувачів переглядають дану тему (1 Гостей і 0 Прихованих Користувачів)
0 Користувачів:

|
Lo-Fi Версія | Поточний час: 13th December 2025 - 20:52 |